GROUP RESOURCES
Summary
Molecular interaction network models describe how different molecules - proteins, small RNAs, any molecules we wish to study - interact with each other, similarly as to how social networks describe how we, humans, interact with one another. Collecting this interaction information helps scientific communities by serving as a knowledge base of all known interactions, and at the same time driving research forward by making it possible to predict interactions which were previously unknown.
Molecular interactions can occur in different “layers”, depending on the qualities of the molecules at hand: for example protein - protein interaction layers describe the direct, physical interactions of proteins, while the transcriptional regulatory layer describes the relationships between transcription factors and their regulated genes. While both of these layers represent interactions in the same organism or cell, they occupy a different lane of information flow, and rarely meet. Multilayered network models aim to connect them, by finding the points where the layers do cross over - such links, referred to as intralayer interactions help us describe molecular interactions networks in a more realistic way, by giving us a more complete picture of the studied system, for example by highlighting how a specific protein - protein interaction is regulated upstream.
Our group has developed and maintained multiple molecular interaction network resources over the past years. Our flagship database, SignaLink, contains manually curated information about signalling pathways and their related interactions, for humans, and for other important model organisms, such as the zebrafish or the fruit fly. Together with the Saez-Rodriguez group we have released OmniPath, a resource combining over 100 other interaction resources of protein-protein interactions, regulatory interactions, cell-cell communication and more. We have developed more specialised resources as well: the AutophagyNet resource contains interaction information specifically focusing on the core autophagy machinery. CytokineLink, our newest resource, introduced a novel interaction type, aiming to describe how cytokines, small proteins orchestrating the proper functioning of the immune system, might affect each other. Our NRF2-ome resource focuses on interaction regarding all things NRF2, a very important transcriptional regulator. SalmoNet is a multi-layered network resource, but built for a non-model organism, the pathogenic bacteria Salmonella enterica. In addition we created an open-source, cloud-based big data platform, called Sherlock, which is providing a gap-filling way for computational biologists to store, convert, query, share and generate biology data, while ultimately streamlining bioinformatics data management.

Figure 1: Summary of our resources and their biological usages.
For full-sized version, please click on the image.
Interaction network resources collate, curate and predict molecular interactions occurring between select entities. Most currently available signalling databases contain only a part of the complex network of intertwining pathways, leaving out key interactions or processes. Multi-layered network resources can bridge this gap, by integrating various levels of communication occurring between the analysed molecules. We have published multiple integrated network resources in the past, all facilitating research in their respective fields (Thomas et al, Frontiers in Genetics, 2021).

For full-sized version, please click on the image.
SignaLink 3
SignaLink3 is our flagship network resource, providing manually curated data on signalling pathways, and integrating other information from several other databases, including, but not limited to, regulation, localisation and disease state (Csabai et al. 2018, 2022). The database contains over 700 000 interactions for Homo sapiens, making it one of the largest integrated signalling network resources. Next to Homo sapiens, SignaLink3 is the only current signalling network resource to provide regulatory information for the model species Caenorhabditis elegans, Danio rerio, and is the largest resource for Drosophila melanogaster.

For full-sized version, please click on the image.
OmniPath
OmniPath is a database of molecular biology prior knowledge (Türei et al. 2021). In our joint work with the Saez-Rodriguez group we combined over 100 resources covering interactions and roles of proteins in inter- and intracellular signalling, as well as transcriptional and post-transcriptional regulation. We added protein complex information and annotations on function, localization, and role in diseases for each protein. The resource is available for human, and via homology translation for mouse and rat. The data are accessible via OmniPath’s web service, a Cytoscape plug-in, and packages in R/Bioconductor and Python, providing access options for computational and experimental scientists. For more information and the full method, please read this paper: Türei et al, Nature Methods, 2016
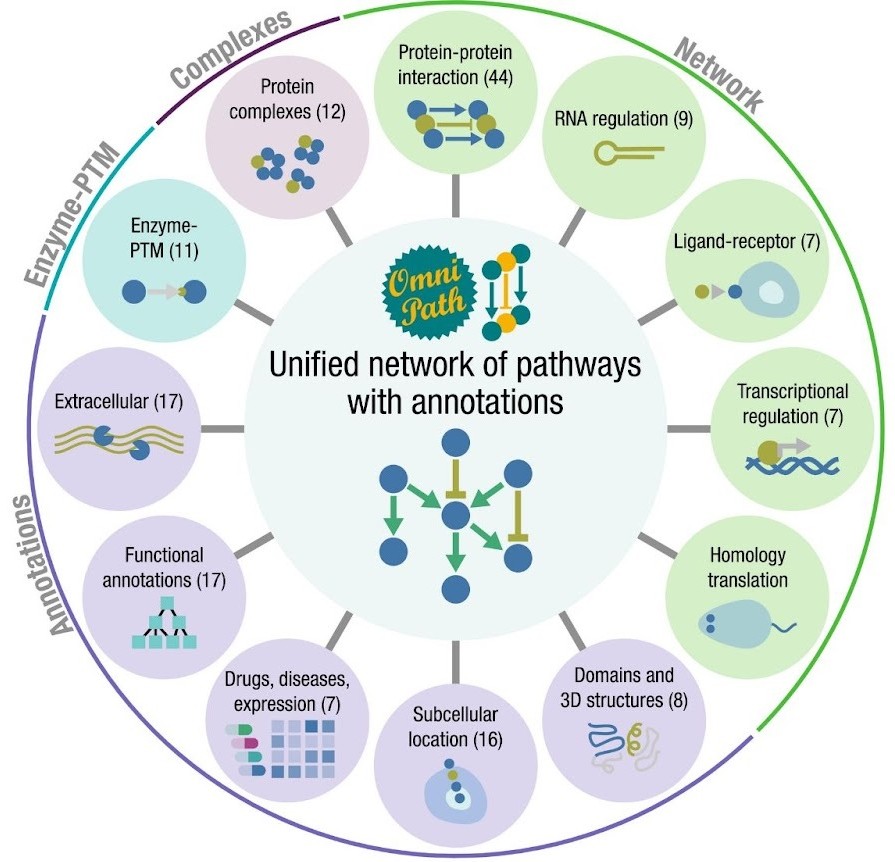
For full-sized version, please click on the image.
AutophagyNet
AutophagyNet is an integrated database for autophagy research, building on the previous prototype of Autophagy Regulatory Network (Türei et al., 2015). AutophagyNet contains updated curation and integration of over 280,000 experimentally verified interactions between core autophagy proteins and their protein, transcriptional and post-transcriptional regulators as well as their potential upstream pathway connections. AutophagyNet provides annotations for each core protein about their role:
- 1) in different types of autophagy (mitophagy, xenophagy, etc.);
- 2) in distinct stages of autophagy (initiation, elongation, termination, etc);
- 3) with subcellular and tissue-specific localization.
These annotations can be used to filter the dataset, providing customizable download options tailored to the user’s needs. The resource is available in various file formats (eg, CSV, BioPAX, PSI-MI), and data can be visualized directly in Cytoscape. The resource is publicly accessible at http://autophagynet.org. Combining networks from AutophagyNet with tissue- or cell-derived, large datasets (e.g. transcriptomics, proteomics), the resource enables (multi-)omics analysis of autophagy regulation.
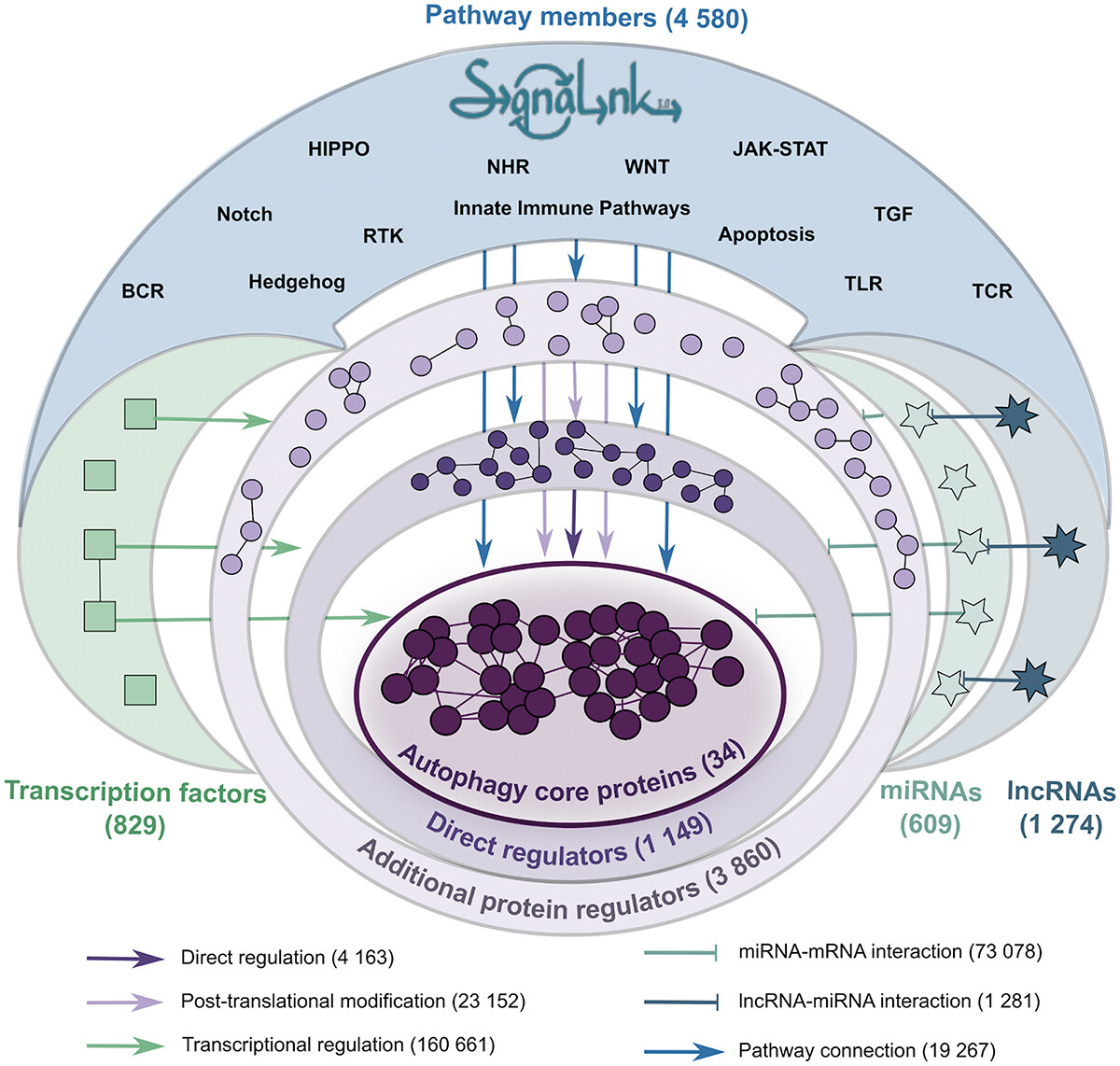
For full-sized version, please click on the image.
CytokineLink
CytokineLink is a specialised resource focussing on how cytokines influence the expression of other cytokines (Olbei et al. 2021). In this effort, we built a communication map between major tissues and blood cells that reveals how cytokine-mediated intercellular networks form during homeostatic conditions. We collated the most prevalent cytokines from the literature and assigned the proteins and their corresponding receptors to source tissue and blood cell types based on enriched consensus RNA-Seq data from the Human Protein Atlas database. To assign more confidence to the interactions, we integrated the literature information on cell–cytokine interactions from two systems of immunology databases, immuneXpresso and ImmunoGlobe. From the collated information, we defined two metanetworks: a cell–cell communication network connected by cytokines; and a cytokine–cytokine interaction network depicting the potential ways in which cytokines can affect the activity of each other.
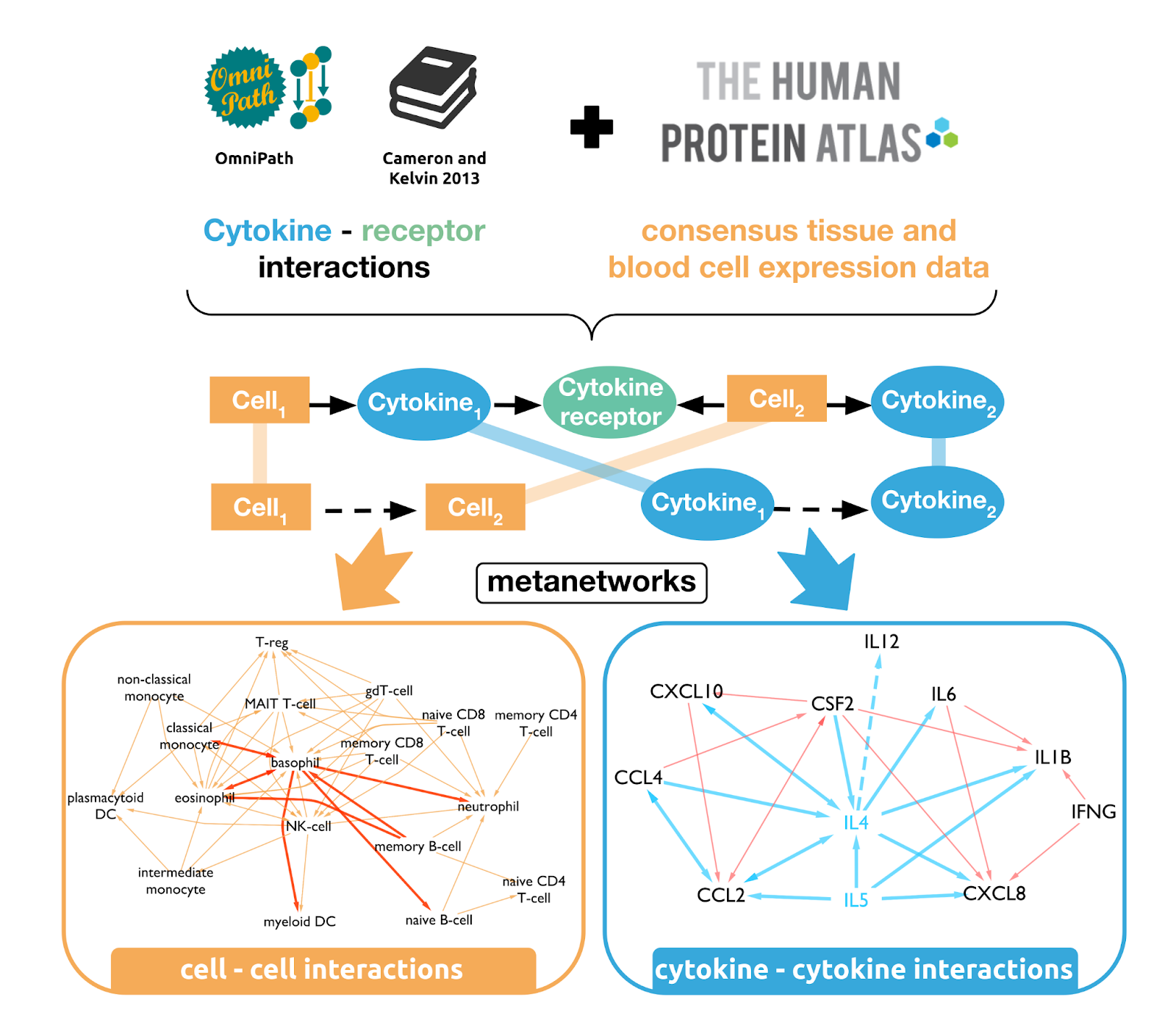
For full-sized version, please click on the image.
NRF2-ome
The NRF2-ome database is an integrated web resource focussing on interactions related to NRF2, the master regulator of oxidative and xenobiotic stress responses, with elevated importance in carcinogenesis, inflammation, and neurodegenerative diseases (Papp et al. 2012; Türei et al. 2013). The database contains manually curated and predicted interactions of NRF2 as well as data from external interaction databases, integrating the NRF2 interactome with NRF2 target genes, NRF2 regulating TFs, and miRNAs.

For full-sized version, please click on the image.
SalmoNet 2
SalmoNet is the first multi-layered interaction resource for Salmonella strains, containing protein-protein, transcriptional regulatory and enzyme-enzyme interactions for all of the individual strains (Métris et al. 2017; Olbei et al. 2019). In its updated version, SalmoNet2 contains multi-layered networks for 20 Salmonella strains, including strains such as S. Typhimurium D23580, an epidemic multidrug-resistant strain leading to invasive non-typhoidal Salmonella Disease (iNTS), and a strain from Salmonella bongori, another species in the Salmonella genus, in total encompassing over 270000 molecular interactions from the studied strains.
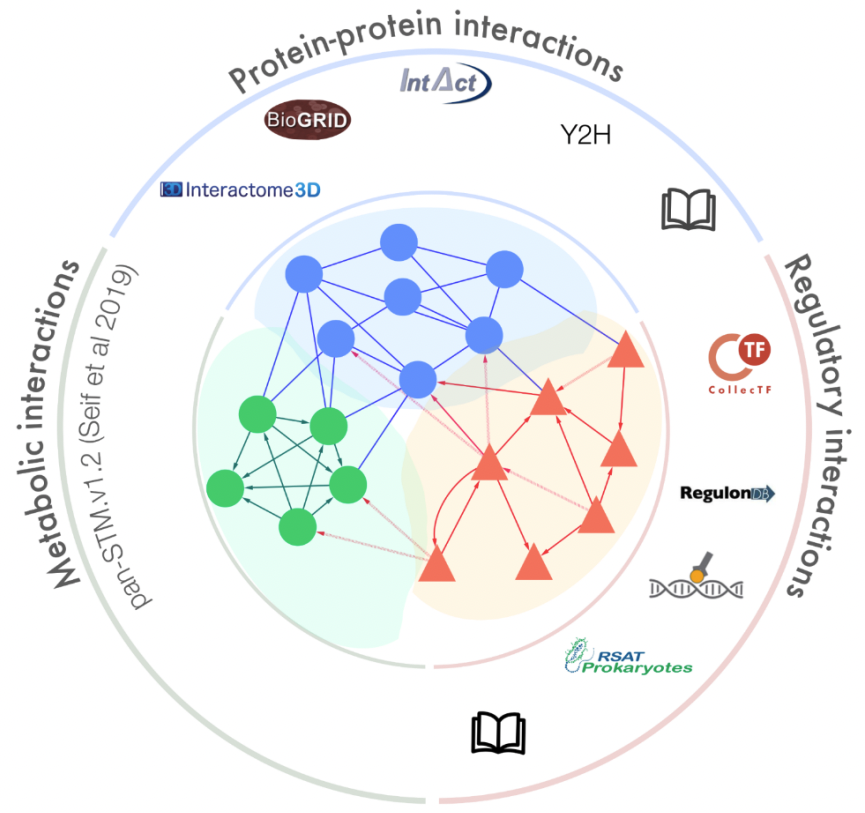
For full-sized version, please click on the image.
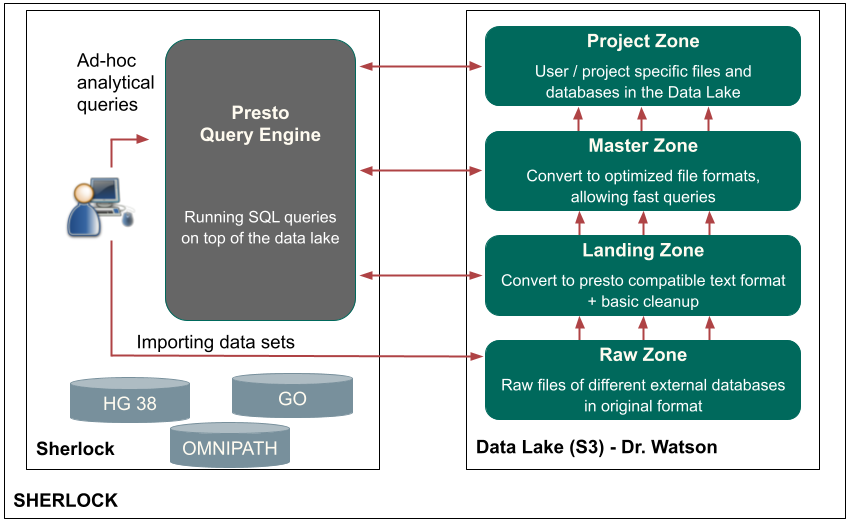
Sherlock platform
Sherlock is an open-source, cloud-based big data platform (GitHub page). Sherlock provides a gap-filling way for computational biologists to store, convert, query, share and generate biology data, while ultimately streamlining bioinformatics data management. The Sherlock platform provides a simple interface to leverage big data technologies, such as Docker, PrestoDB or HIVE Metastore. Sherlock is designed to analyze, process, query and extract the information from extremely complex and large data sets. Furthermore, Sherlock is capable of handling different structured data (interaction, localization, or genomic sequence) from several sources and converting them to a common optimized storage format. This format facilitates Sherlock’s ability to quickly and easily execute distributed analytical queries on extremely large data files. The Sherlock platform is freely available on github, and contains specific loader scripts for structured data sources of genomics, interaction and expression databases. With these loader scripts, users are able to easily and quickly create and work with the specific file formats. For further details, please check our publication about Sherlock: (Bohár B. et al, 2021). More details here.
For full-sized version, please click on the image.