Project – Network Medicine Approaches in IBD
Inflammatory Bowel Disease (IBD) is a disorder of the human gastrointestinal tract characterised by inflammation of the gut mucosal layer and dysbiosis of the microbiome. IBD can broadly be classified into two diseases - Ulcerative Colitis (UC) and Crohn’s Disease (CD) - which are increasingly prevalent in the global population. However, IBD is a complex disease influenced by a variety of extrinsic and intrinsic factors such as genetics, microbiome, lifestyle (diet, smoking etc), xenobiotic exposures etc. In the Korcsmaros group, we aim to understand IBD using multiple different angles and various omics techniques as input data sources (Figure 1).
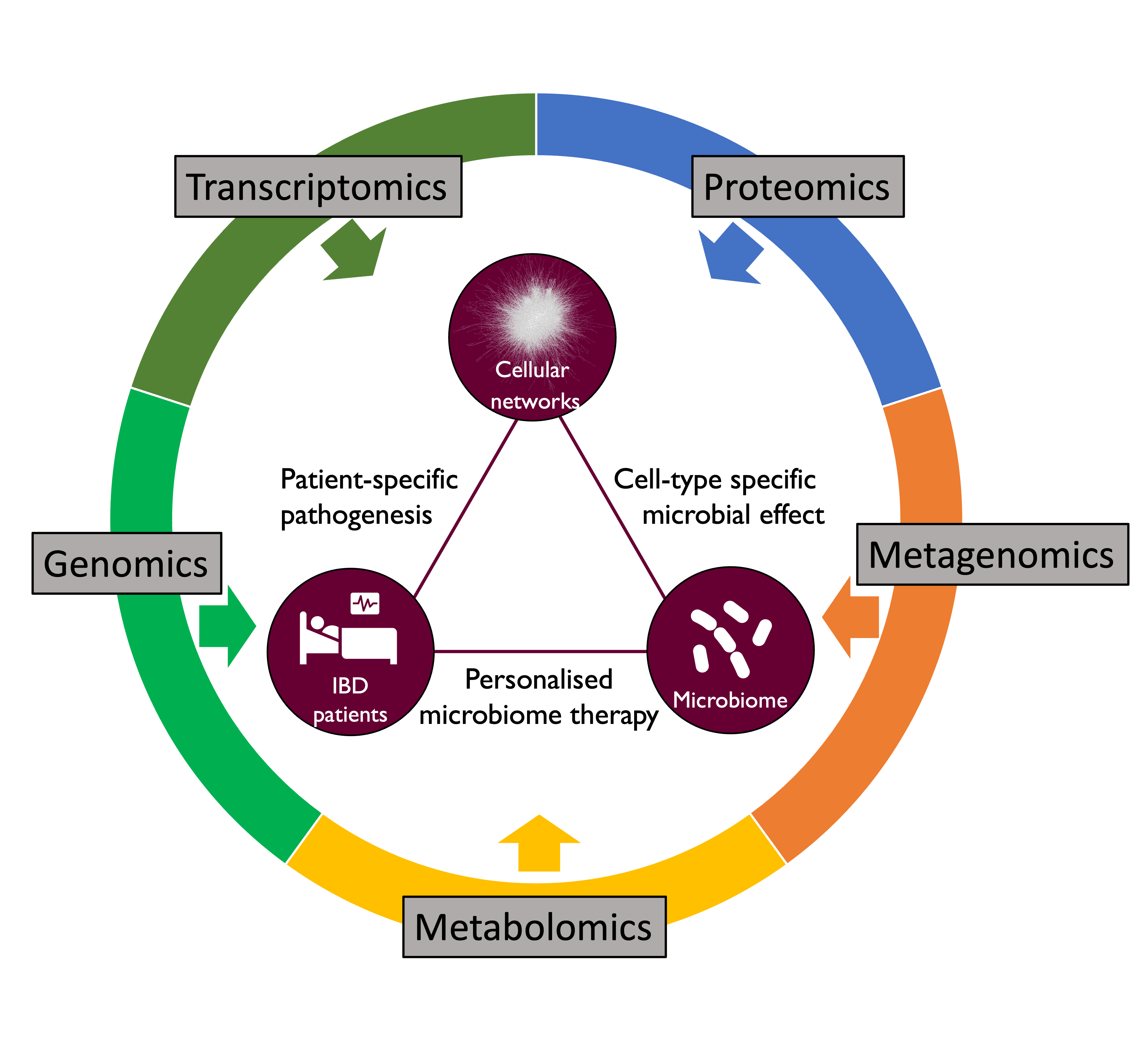
Figure 1: Our integrative research approaches to study IBD.
For full-sized version, please click on the image.
Precision medicine in IBD
To investigate patient-specific pathogenesis in IBD, we collaborate with Séverine Vermeire’s IBD team from Leuven and work in the UK IBD Genetic Consortia. We use patient-specific genomics and transcriptomics information from these collaborators to decipher how patient-specific genomic background contributes to IBD. Using known IBD single nucleotide polymorphisms (SNPs), we have developed the iSNP, precision medicine pipeline. iSNP is able to identify regulatory SNPs in the genome, and integrate regulatory and physical interaction data to improve our understanding of IBD pathogenesis (Brooks et al, Nature Communications, 2022).
Most of the known SNPs in IBD are in the regulatory, non-coding regions of the genome. Interpreting the effect of non-coding SNPs is challenging. To address this issue, we were able to map many of them to various transcription factor binding site and miRNA target site regions. Then we are investigating both upstream and downstream processes. In other words, how the disease-associated SNPs effect which upstream regulator is effecting the expression of a gene (ie, change in the incoming pathway) as well as how the altered expression of a gene due to a SNP is effecting downstream signalling processes and contributes to the detected disease phenotype (ie, change in the regulatory landscape or transcriptome).
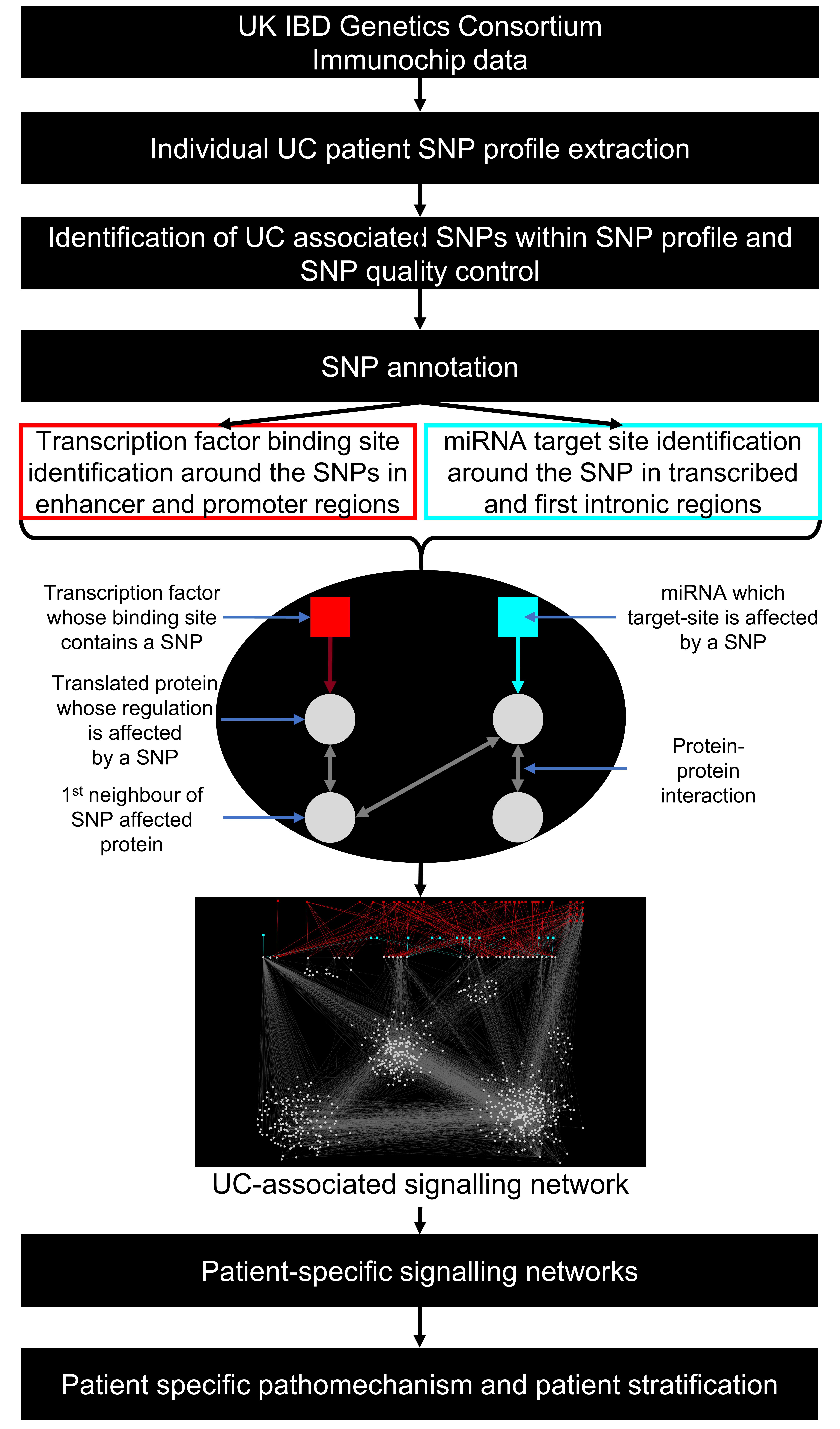
Figure 2: The workflow of the iSNP pipeline (Brooks et al, Nature Communications, 2022)
For full-sized version, please click on the image.
Intercellular interactions in IBD
In inflammatory bowel diseases, various cytokines have an enormous role both in the pathogenesis of the disease and as targets for treating the condition. We built a database of cytokine-specific interactions called CytokineLink. Using cytokine-cytokine interactions between the various cells in the mucosal immune system could show how and what interactions are affected in IBD. Analysing epithelial-immune cell interactions is crucial to understand the molecular background of IBD. Collaborative projects led by Nick Powell’s group at Imperial College highlighted the role of cytokines in the healthy gut and in IBD conditions (Pavlidis et al, Cell Reports, 2022; Pavlidis et al, Nature Communications, 2022). We combined experimental data from organoids with network biology approaches to model the effect of five cytokines (IL13, IL17A, TNFa, IFNγ and IL22). The analysis helped to reveal signalling bottlenecks in cytokine response and highlighted their translational potential as predictive biomarkers for therapeutic response in intestinal inflammation.

Figure 3: The integrated workflow using transcriptomics data of organoids exposed to various cytokines, and network biology approaches to identify key profiles that are present in patient cohort data. Image credit: Leila Gul and Pavlidis et al, Cell Reports, 2022
For full-sized version, please click on the image.
The recent abundance of single cell RNA-seq genomics techniques made it possible to build intercellular networks to understand inflammatory bowel disease pathogenesis. We leverage these data sources to map the intercellular landscape of IBD. Using single cell RNA-seq data such as Martin et al, Smillie et al, or the Gut Cell Atlas, and network resources such as OmniPath, we model the interactions between cells,a nd how they change between healthy and disease, or inflamed vs. uninflamed conditions (in the same patient).
Towards personalised microbial therapy in IBD
IBD as a multifactorial disease has both genetic and environmental factors. One of the main environmental factors is the microbiome, and faecal microbiota transplantation has been suggested as a potential treatment option for IBD. However, the exact details of how various microbes could improve gut health and in which patient they could be effective are yet to be discovered. The aim of our group is to improve our understanding on which microbes could be the most effective treatment for which patient. In collaboration with the groups of Falk Hildebrand and Simon Carding in the Quadram Institute (Norwich, UK) we are investigating the interactions between patients and microbes using metagenomics, proteomics, and single-cell sequencing.
We have developed the Microbiolink pipeline (Tahila et al, Cells, 2020) to decipher potential host-microbe cross-talks. Using single cell RNA-seq data, we showed which cells could interact with which microbial proteins, and in which condition (inflamed or uninflamed). This study pointed out the importance of cell-type and condition-specific analyses as beneficial microbes may have a well-defined time and target cells (Gul et al, Journal of Extracellular Vesicles, 2022).
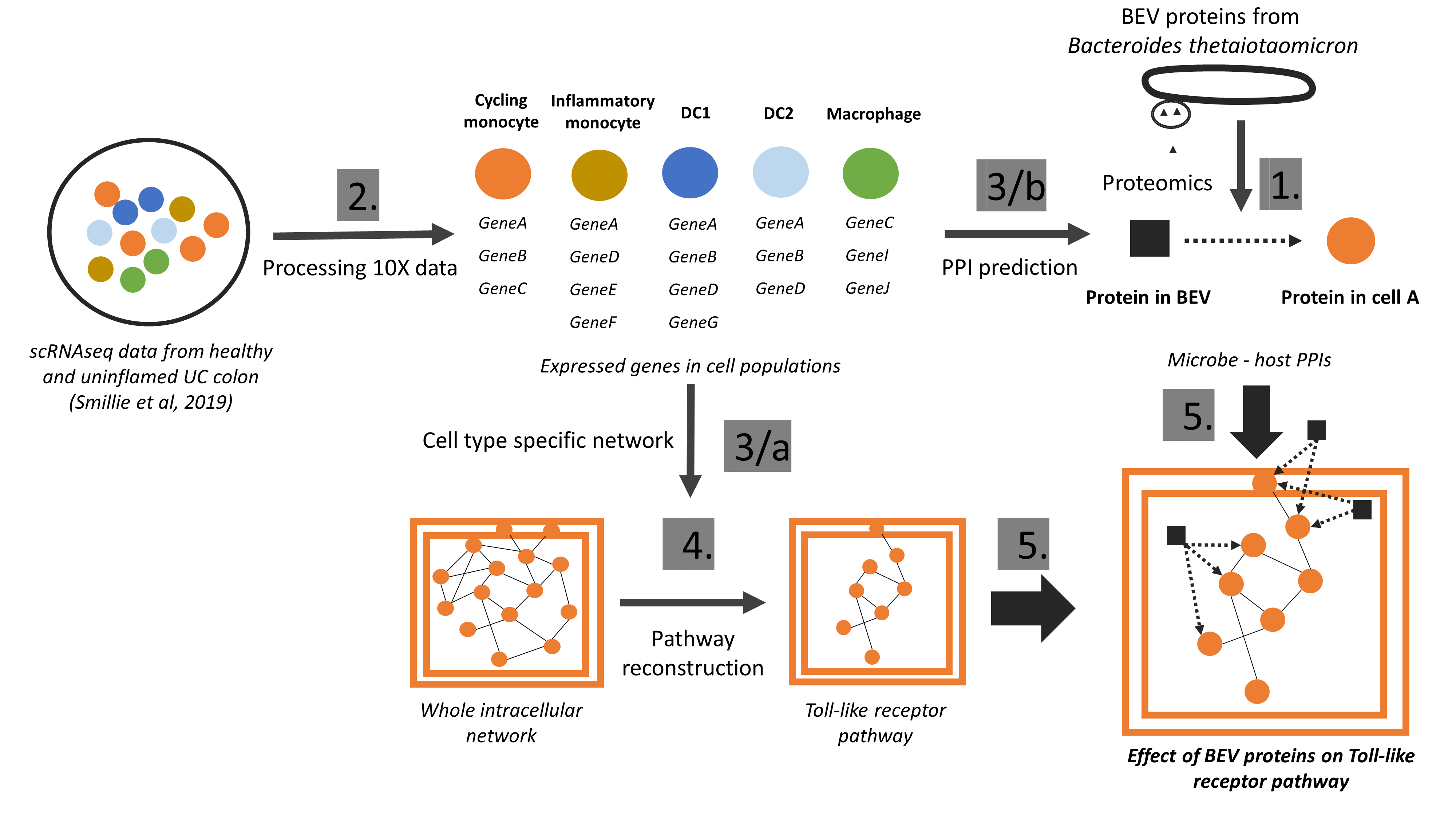
Figure 4: The workflow of the MicrobioLink pipeline using single-cell data on hsot cells and proteomic data from microbes to identify host-microbe interactions and their downstream effect. (Gul et al, Journal of Extracellular Vesicles, 2022)
For full-sized version, please click on the image.