Korcsmaros Group
Network Medicine & Organoids
Introduction
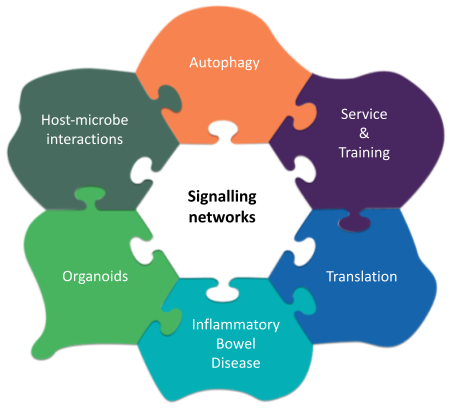
 The Korcsmaros Group combines computational approaches (network biology and AI) and experimental technologies (organoids and multi-omics) to study complex systems in the gut. Our goal is to understand biological systems related to gut homeostasis and to facilitate precision medicine and personalised microbial therapies in the field of inflammatory bowel disease (IBD).
Our vision is that with the combination of systems biology and precision medicine resources, we are going to understand better the patient-specific nature of host-microbe interactions in the gut, and identify effective therapeutical options preventing the onset of IBD or relapse. To achieve this vision, our aim is to work on the multi-omics analysis of gut organoids to create computational network models of key intestinal cells interacting with other host cells or with microbial products, and then validate the predictions in a patient-specific organoid testing system.
The Korcsmaros Group combines computational approaches (network biology and AI) and experimental technologies (organoids and multi-omics) to study complex systems in the gut. Our goal is to understand biological systems related to gut homeostasis and to facilitate precision medicine and personalised microbial therapies in the field of inflammatory bowel disease (IBD).
Our vision is that with the combination of systems biology and precision medicine resources, we are going to understand better the patient-specific nature of host-microbe interactions in the gut, and identify effective therapeutical options preventing the onset of IBD or relapse. To achieve this vision, our aim is to work on the multi-omics analysis of gut organoids to create computational network models of key intestinal cells interacting with other host cells or with microbial products, and then validate the predictions in a patient-specific organoid testing system.
Past Projects
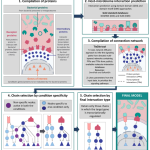
 We developed gap-filling network biology resources on signalling pathways and autophagy regulation (such as SignaLink, AutophagyNet, CytokineLink, SalmoNet) that we provide for the community through Cyverse UK. We also developed multiple computational workflows and software platforms to carry out automated analysis of big data in multi-omics and precision medicine (including iSNP, MicrobioLink, and Sherlock). In the past, we worked with various experimental model systems, including C. elegans, human cell lines and mouse organoids, and developed new protocols to investigate gut organoids and host-microbe interactions.
We developed gap-filling network biology resources on signalling pathways and autophagy regulation (such as SignaLink, AutophagyNet, CytokineLink, SalmoNet) that we provide for the community through Cyverse UK. We also developed multiple computational workflows and software platforms to carry out automated analysis of big data in multi-omics and precision medicine (including iSNP, MicrobioLink, and Sherlock). In the past, we worked with various experimental model systems, including C. elegans, human cell lines and mouse organoids, and developed new protocols to investigate gut organoids and host-microbe interactions.
These resources and methods provided the following significant biological insights:
- the hidden importance of first neighbours (protein interactors) of mutated proteins in cancer and in IBD that allowed us to define novel biomarkers and drug repurposing options (Modos et al, NPJ System Biology and Applications, 2017; Brooks et al, Nature Communications, 2022);
- a complex evolutionary interplay between bacterial and host autophagy proteins that pointed out parallel evolutionary mechanisms, explanation why certain infections are higher in IBD, and potentially novel antimicrobial drug targets (Sudhakar et al, Autophagy, 2019; Demeter et al, Frontiers in Cellular and Infection Microbiology, 2022)
- a new mechanism on how autophagy dysfunction contributes to IBD, and what processes may also be affected upon treating a patient with autophagy modulating drug (Jones et al, Disease Models & Mechanisms, 2019);
- a cell- and condition-specific role of bacterial proteins to affect key cell types in the gut, pointing out how bacterial extracellular vesicles can be used to support IBD treatment (Gul et al, Journal of Extreacellular Vesicles, 2022);
- multiple cytokine-specific communication channels between intestinal epithelial cells and immune cells that contribute to the generation of inflammation and cytokine storm upon infection and IBD (Poletti et al, NPJ Systems Biology and Applications, 2022; Pavlidis et al, Cell Reports, 2022; Pavlidis et al, Nature Communications, 2022).
We have summarized in multiple review papers the key aspects and opportunities of the main methodologies we use, including Network Approaches, Organoids, Big Data and IBD, Big Data and autophagy, studying host-microbe interactions with computational and with experimental approaches.
Current and Future Projects
Intestinal homeostasis is maintained by multiple cell types, including Paneth cells and goblet cells, producing antimicrobial peptides and the mucus layer, respectively, and shaping the microbiome and regulating how our cells are interacting with it. One of the key cellular processes in contributing to these functions is autophagy. Building on our previous achievements and developed approaches, our multidisciplinary group aims to investigate how autophagy is regulated in Paneth and goblet cells upon exposure to external stimuli from other host cells or microbes, and to decipher how host mutations often associated with IBD affect these intercellular and inter-kingdom cross-talks. This approach provides a holistic, systems-level view of IBD-related processes in the gut, and will complement existing programs focusing on immune-therapies against IBD (ie., Crohn’s disease (CD) and ulcerative colitis (UC)). Another key approach we consider is the patient-specific nature of these observations. We plan to understand the differences between CD/UC patient subpopulations. Characterising patient group-specific alterations in interactions between host cells or between host cells and the microbiome; with a particular focus on interactions related to autophagy will lead us to provide the foundations of novel microbial and drug therapies. With these approaches we are addressing the complex questions of how key cells important for the intestinal homeostasis are regulated by other host cells or by gut microbes, how this regulation is changing upon IBD, and how we can use the identified knowledge to suggest novel therapeutic approaches for improving gut health.
We are collaborating with multiple groups to achieve these objectives including groups in our host institute at Department of Metabolism Digestion and Reproduction, Faculty of Medicine, Imperial College London as well as in our affiliated institute in the Quadram Institute. In particular, we have strategic collaborations with the following groups:
- Nick Powell lab (Imperial College London, UK)
- Séverine Vermeire and Bram Verstock (IBD Leuven, Belgium)
- Julio Saez-Rodriguez group (University of Heidelberg)
- Paul Wilmes group (University of Luxembourg)
- Lindsay Hall Lab (Technical University Munich, Germany
- Orsolya Kapuy (Semmelweis University, Hungary)
- Ioannis Nezis (University of Warwick, UK)
- Tom Wileman (Quadram Institute, Norwich, UK)
- Simon Carding (Quadram Institute, Norwich, UK)
- Falk Hildebrand (Quadram Institute, Norwich, UK)
- Cynthia Whitchurch (Quadram Institute, Norwich, UK)
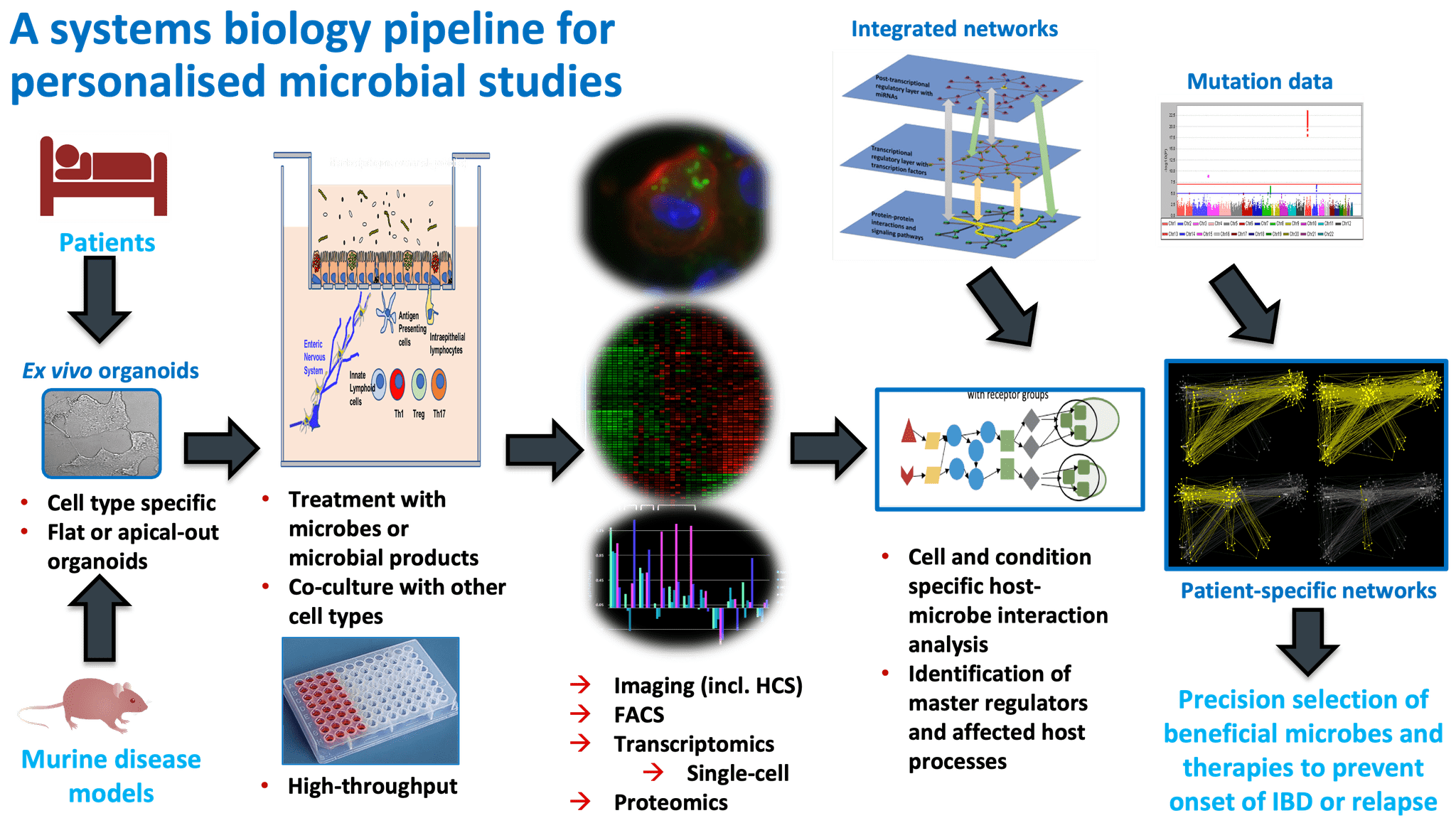
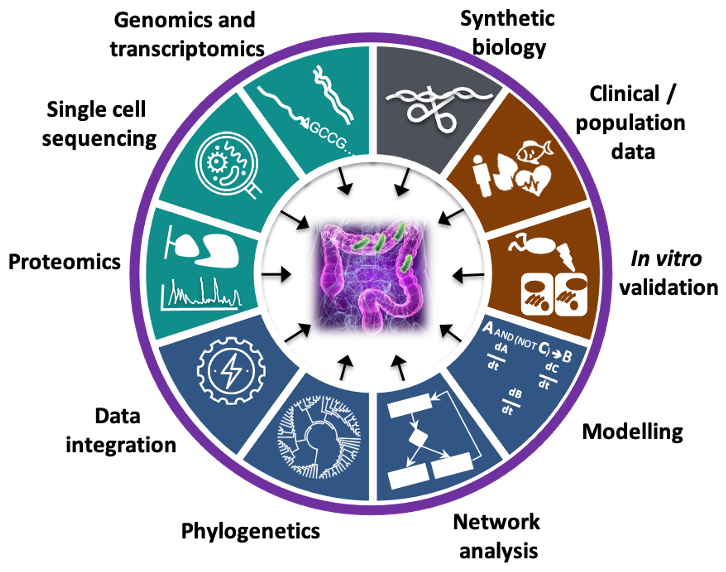
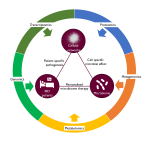
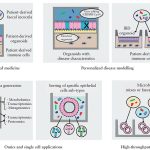
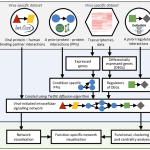
Overall pipeline of our methods.
For full-sized version, please click on the image.
Our work has been supported by the NIHR Imperial BRC and institute strategic grants from the UKRI-BBSRC to the Quadram Institute and the Earlham Institute
Resources
Projects
Affiliations
 – Faculty of Medicine, Imperial College, London, United Kingdom
– Faculty of Medicine, Imperial College, London, United Kingdom
 – Earlham Institute, Norwich Research Park, Norwich, United Kingdom
– Earlham Institute, Norwich Research Park, Norwich, United Kingdom
 – Gut Microbes and Health Programme, Quadram Institute Bioscience, Norwich Research Park, Norwich, United Kingdom
– Gut Microbes and Health Programme, Quadram Institute Bioscience, Norwich Research Park, Norwich, United Kingdom
 – Department of Genetics, Eotvos Lorand University, Budapest, Hungary
– Department of Genetics, Eotvos Lorand University, Budapest, Hungary